Introduction: Recent studies using next generation sequencing have revealed recurrent mutated genes in CLL are associated with clinical outcome. We assessed the baseline mutational profile of 771 patients from FLAIR treated with either fludarabine, cyclophosphamide and rituximab (FCR) or ibrutinib and rituximab (IR). The prognostic impact of individual gene mutations on disease progression was investigated.
Method: FLAIR is a phase III, multicentre, randomised, controlled, open, parallel group trial for previously untreated CLL requiring therapy according to IWCLL criteria. Patients with >20% chromosome 17p deletion were excluded. Patients were randomised to FCR or IR. Somatic hypermutation (SHM) status was determined by PCR amplification of IGHV-IGHD-IGHJ gene rearrangements using IGHV leader/FR1 primers. Bidirectional Sanger sequencing was analysed using IMGT V-Quest and the ARResT/AssignSubsets tool. Extracted DNA was sequenced using an Illumina MiSeq and analysed using an in-house pipeline. Amplicon based targeted sequencing of 33 recurrently mutated genes in lymphoid malignancies were performed in parallel. Detected variants were reported down to minimum variant allele fractions of 3-5% and coverage of 100X. Low level variants were confirmed by repeat sequencing.
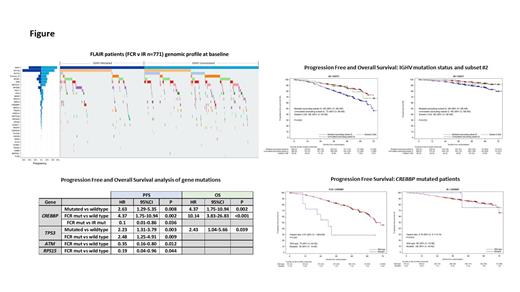
Results: 771 patients were randomly assigned to receive FCR (385) or IR (386). IGHV gene SHM status was available for 728/771 (94.4%): 388 (53.3%) IGHV unmutated (>98% of nucleotide identity to germline), 294 (40.4%) IGHV mutated and 46 (6.3%) stereotyped subset #2 (n=28 IGHV mutated, n=18 IGHV unmutated). Gene mutations were assessed in 767/771 (99.5%) patients at baseline and mutations (>1) were detected in 480/767 (62.6%). Mutation frequencies ranged from 0.1-18.8% with mutations in SF3B1 (18.8%), ATM (14.5%), NOTCH1 (10.0%), MYD88 (6.1%), POT1 (5.7%), TP53 (5.0%), BRAF (4.4%) and RPS15 (4.0%) being the most frequent. No detectable mutations at baseline were found in 287/767 (37.4%) and there was no significant difference in PFS or OS compared to patients with >1 mutation.
A significantly shorter PFS and OS was observed for TP53 mutated patients compared to wildtype (HR 2.23 [95% CI 1.31-3.79]; p=0.003 and HR 2.43 [95% CI 1.04-5.66]; p=0.039 respectively). When further subdivided by treatment, a trend for shorter PFS was observed in TP53 mutated patients in both arms compared to wildtype, but significance was only achieved for FCR (HR 2.48 [95% CI 1.25-4.91]; p=0.009) and not IR (HR 2.27 [95% CI 0.97-5.30]; p=0.059). A significantly shorter PFS was observed for FCR compared to IR in patients with ATM mutations at baseline (HR 0.35 [95% CI 0.16-0.80]; p=0.012) and RSP15 (HR 0.19 [95% CI 0.04-0.96]; p=0.044). A trend for shorter PFS was also observed in NOTCH1 mutated patients treated with FCR however, this was not found to be significant when compared to wildtype or NOTCH1 mutated IR patients.
CREBBP mutation at baseline (n=19; 2.5%) was associated with shorter PFS (HR 2.63 [95% CI 1.29-5.35]; p=0.008) and OS (HR 4.37 [95% CI 1.75-10.94]; p=0.002) compared to wild type (Fig). When further subdivided by treatment arm, shorter PFS and OS was also observed for CREBBP mutated patients treated with FCR compared to wildtype (n=11; HR 4.37 [95% CI 1.75-10.94]; p=0.002 and HR 10.14 [95% CI 3.83-26.83]; p<0.001 respectively). A similar trend was not observed with IR (n=8; PFS HR 0.79 [CI 0.11-5.74]; p=0.819 and OS HR NE [95% CI NE-NE]; p=0.988). CREBBP mutated patients treated with FCR had a shorter PFS compared to IR (HR 0.1 [95% CI 0.01-0.86]; p=0.036).
The PFS and OS for IGHV subset #2 was similar to IGHV mutated patients (Fig). In FCR treated patients there was no difference between mutated (n=13) and unmutated subset #2 (n=7) but with IR there was a trend that IGHV mutated subset #2 patients (n=15) had better PFS and OS than their IGHV unmutated counterparts.
Conclusions: Our study confirms that TP53 mutations result in shorter PFS and OS in CLL. We demonstrate improved PFS when patients with ATM or RPS15 mutations are treated with IR compared to FCR. Mutations in the CREBBP gene have recently been described as a novel candidate driver in CLL. We report a significant improvement in disease progression in CREBBP mutated patients treated with IR when compared to FCR. In contrast to previous studies in frontline CLL IGHV subset #2 patients had a similar outcome to good risk IGHV mutated patients.
Disclosures
Rawstron:Abbvie: Honoraria, Research Funding; Janssen: Honoraria, Research Funding; Pharmacyclics: Consultancy, Research Funding; Beigene: Consultancy, Honoraria, Research Funding; Gilead: Research Funding. Cairns:Takeda: Research Funding; Celgene BMS: Honoraria, Research Funding; Sanofi: Research Funding; Amgen: Research Funding; Janssen: Honoraria. Hockaday:Abbvie: Speakers Bureau. Bloor:Abbvie: Consultancy, Honoraria, Speakers Bureau; Gilead, Janssen: Honoraria. Munir:Janssen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Abbvie: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; AstraZeneca: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Roche: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Sobi: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Alexion: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; BeiGene: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau. Hillmen:Apellis Pharmaceuticals: Current Employment, Current equity holder in publicly-traded company.